Intrinsic disorder in proteins has fascinated me ever since 1997, when Kevin Plaxco asked me to co-author a News & Views piece (1) on what was then an emerging topic. By now it is an established scientific phenomenon feeding a whole research community, so I even had the opportunity to attend a conference about it a few years ago, and write a feature for Chemistry World among other articles.
We now know that intrinsically disordered proteins play an important role in nature. Quite a few of them work in molecular recognition and can achieve specific binding by “folding around” their target. Others are medically relevant. For instance, there are disordered domains (an oxymoron for the protein folding crowd to chuckle or argue about) in virus proteins and in transcription factors that are important targets for cancer drugs.
If nature can find use for disordered sequences, maybe scientists can also use them in molecular design? Kevin’s group at the University of California at Santa Barbara has now demonstrated an intriguing approach in which disordered sequences (of DNA, this time) can make a receptor more cooperative, meaning more likely to bind a second molecule once it has bound the first (2). The best known natural example of molecular cooperativity is the binding of oxygen to haemoglobin in our red blood cells – it can carry up to four molecules, and each position filled increases the affinity of the remaining ones. The attraction, for haemoglobin as for biotechnologists, is that cooperative binding has a much sharper transition, switching from all empty to all full in a narrower range of concentrations than a non-cooperative receptor would.
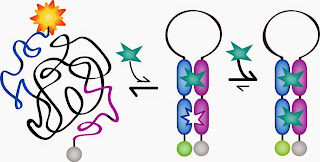
But how do you force a receptor to be cooperative if it isn’t naturally inclined to do this? What first author Anna Simon and colleagues in the Plaxco lab did was to cut the receptor (a DNA aptamer in this work, but it should in principle be possible with proteins as well) in two halves, then duplicate each half. If you think of a complete working receptor as a pair of robotic hands that can grab a ball, they glued two left hands together and two right hands, but a connected pair of left and right was needed to carry out the desired function. They then connected the ends of the left and right construct with a DNA sequence that prefers to be disordered.
Bringing one pair of robot hands together to grab one ball comes at a cost, as the disordered DNA linker loses entropy (i.e. opportunities to adopt many random conformations) when its two ends are brought closely together. Once the first ball is firmly grabbed, however, and this entropic fee has been paid, the second pair of hands is suitably arranged in close proximity and ready to grab the second ball without having to pay any entropic costs for that. Thus, as in haemoglobin, the second binding event is much more favourable than the first.
Image: Anna Simon / ref. (2)
Simon et al. tried this out with three different DNA receptors, from a primitive one binding mercury ions to a sophisticated aptamers for the molecules cocaine and doxorubicin, and found that all showed some cooperativity, and one receptor, the one for doxorubicine, gave results within the error margins of the values that theory predicts for perfect cooperativity.
Seeing this works with all three DNA receptors tested, it should also work with others and could also be transferred to proteins. In fact, a recent paper suggests that nature also uses this trick in proteins already (3). Then it could be expanded to more than two binding sites, and it would be good to have high-resolution structures of these constructs to analyse their function in detail. The application of disorder in molecular engineering may be a whole new research field that has just been born.
references
(1) K. W. Plaxco and M. Groß, Nature 1997, 386, 657.
(2) A. J. Simon et al, Proc. Natl. Acad. Sci. USA 2014, DOI: 10.1073/pnas.1410796111
(3) A. C. Ferreon et al., Nature 2013, 498, 390.














No comments:
Post a Comment